Breadcrumb
- Home
- RPA Properties
RPA Properties
REPLICATION PROTEIN A PROPERTIES
Schematic of human RPA
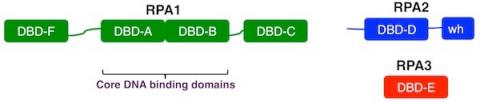
All eukaryotes have trimeric single-strand DNA-binding proteins homologous to human RPA. In most organisms the subunits of RPA are approximately 70, 32, and 14 kDa and are referred to as RPA1, RPA2, and RPA3, respectively. Each of the RPA subunits contains at least one DNA-binding domain (DBD) designated with letters A-F. The DBDs are structurally related, all have oligonucleotide-oligosaccharide-binding (OB) folds.
RPA1 (green) contains four OB-folds. The central two OB‑folds (DBD-A and DBD-B) are both necessary and sufficient for high affinity ssDNA-binding and are referred to as the “DNA-binding core”. DBD-F, at the N-terminus, is needed for efficient binding to partially duplex DNA and participates in checkpoint activation. DBD-F is also required for regulation by phosphorylation, and the cellular damage response. DBD-C, at the C-terminus, interacts with DNA, is required for trimer formation, and is needed for recognition of some types of DNA damage.
RPA2 (blue) contains a central OB‑fold (DBD‑D), an N-terminal phosphorylation domain and a C-terminal winged helix domain. RPA2 interacts with DNA and multiple proteins. Protein interactions are predominantly mediated through the winged helix (WH) domain. The N-terminal domain of RPA2 is also multiply phosphorylated after DNA damage. Phosphorylation of RPA2 modulates RPA activity and is involved in cellular recovery after DNA damage.
RPA3 is composed exclusively of an OB‑fold (DBD‑E). RPA3 interacts weakly with DNA and is required for formation of the RPA complex.
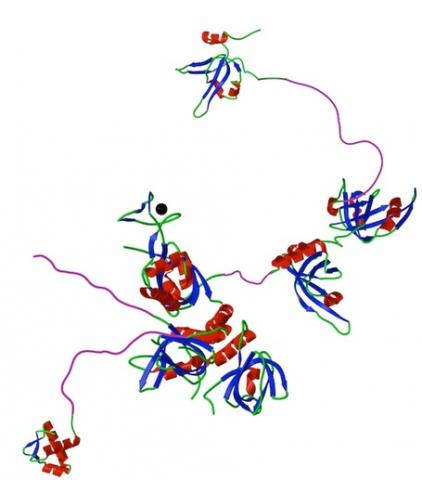
Hypothetical composite model of RPA with structural domains flexible linkers shown to scale. Ribbon structures of DBD F , DBD A and B, the trimerization core (DBDs C, D and E) and the winged helix at the C-terminus of RPA2. Flexible N-terminal phosphorylation domain of RPA32 (Phos) and linker regions are shown to scale in an arbitrary, extended conformation (purple). Zinc ion associated with DBD C is shown as a black circle.
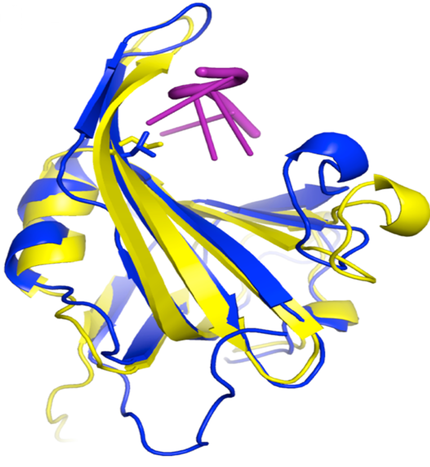
Structure of DNA binding domain A human (blue) and yeast (yellow) RPA1 bound to DNA with the position of the L221 side chain shown extending toward DNA. Mutation of L221 to proline causes inactivates RPA and causes increased genomic instability and cancer. From Hass et al 2010 Mol. Cancer Res. 8, 1017.
© MARC WOLD 2011